Research
Linking Gene Regulation to Metabolism
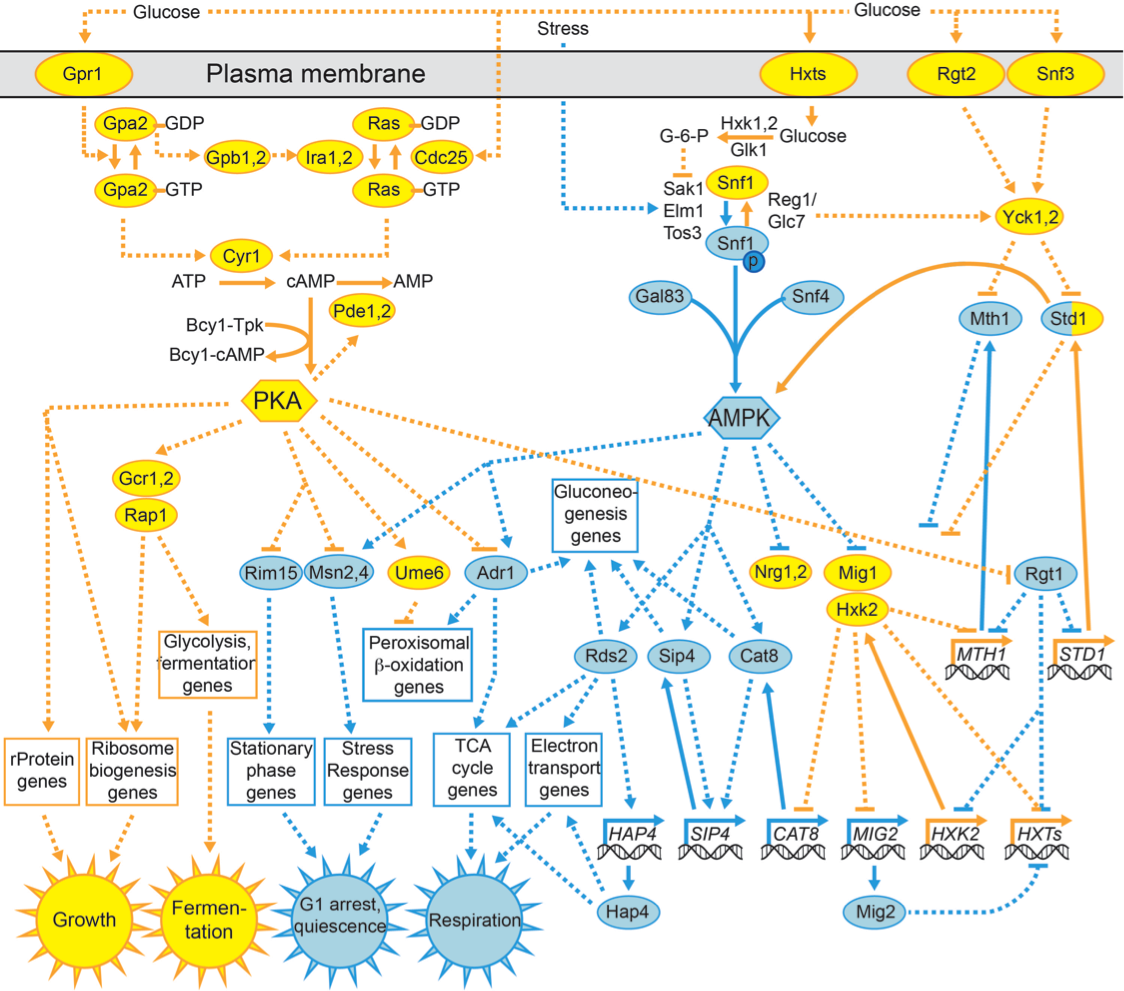
Partial sketch of the overlapping PKA and AMPK pathways. Gold: more active in higher glucose concentrations. Blue: more active in low glucose or no glucose. MTH1 and several HXT (hexose transporter) genes are repressed in both high and low glucose but are expressed at intermediate concentrations. Dotted lines with arrow heads: activation; dotted lines with flat heads: repression; solid lines: covalent or non-covalent transformation.
Many health problems result from, or are dependent on, disregulation of central metabolism. For example, aggressive cancer cells shunt carbon away from aerobic respiration toward anaerobic energy production and biomass. The three master regulators of carbon fate are conserved from yeast to man: Protein Kinase A (PKA), AMP Activated Protein Kinase (AMPK), and Target of Rapamycin (TOR). The long term goal of this project is to understand how the responsibility for carbon fate is divided among these three master regulators and the transcription factors (TFs) downstream of them in mammalian cells. As a first step, we are studying the regulation of carbon fate by AMPK and PKA in the model yeast Saccharomyces cerevisiae. Quite a few of the DNA-binding TFs that serve as end effectors AMPK and PKA are known. Each TF regulates genes encoding enzymes in several pathways and each pathway is regulated by several TFs. Within any given pathway, some genes are regulated by a single known TF, some by several, and some by none. The outcome of this proposal will be a significant step toward understanding how AMPK and PKA regulate carbon fate by coordinating the activities of these end effector TFs.
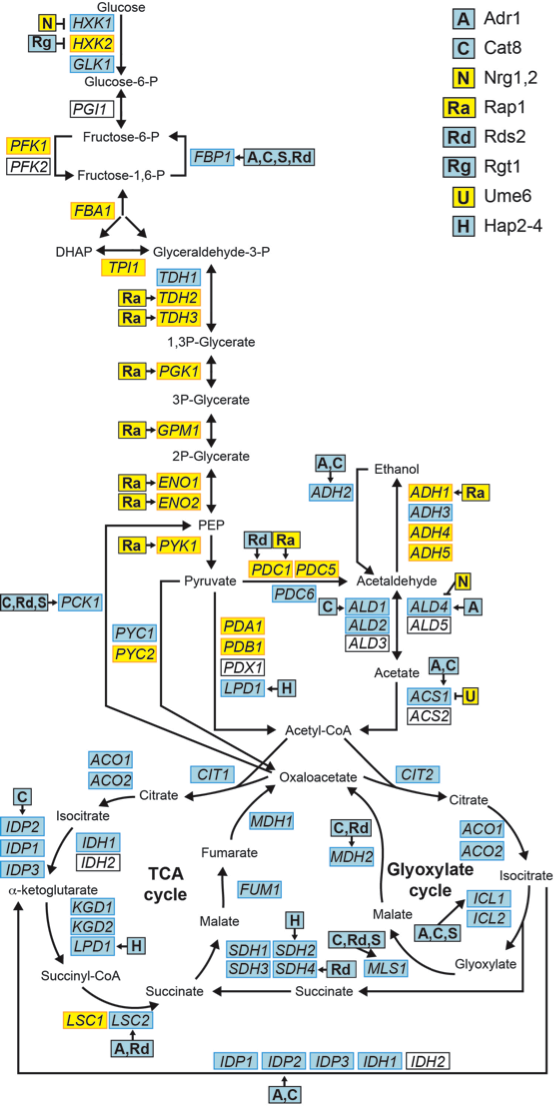
Metabolic pathways annotated with enzymes that catalyze each reaction and transcription factors that regulate each enzyme.
Aim 1. Elucidate the influence of AMPK and PKA on gene expression and carbon fate. To achieve this aim, we will grow yeast with growth-limiting glucose supplies (in which AMPK is active and PKA is not) or excess glucose supplies (in which PKA is active and AMPK is not), monitor carbon fate, and carry out gene expression profiling. We will also carry out these experiments with mutant strains in which we can control the activation level of the AMPK and PKA independently of glucose availability.
Aim 2. Quantify the role of each effector TF in mediating the influence of AMPK and PKA on gene expression and on carbon fate. To achieve this aim, we will carry out experiments like those of Aim 1 using mutants in which we can control AMPK and PKA activation independently and one of 12 downstream TFs has been deleted.
Aim 3. Build a quantitative model linking PKA and AMPK to metabolic outcomes via effector TFs. To achieve this aim, we will construct a quantitative model of gene regulation downstream of AMPK and PKA. We will also estimate metabolic fluxes and construct a model of the effect of enzyme gene expression on metabolic fluxes and hence carbon fate. Taken together, these two models will make quantitative predictions about how both gene expression and carbon fate would be affected by interventions in the regulatory system. Finally, we will test these predictions by using strains in which individual enzymes have been deleted or pairs of TFs have been deleted.
Current Students:
 |
Mike ToomeyWebsite: https://deweezz.com/idn-poker/ |
Collaborators:
 |
Tamara Doering, Ph.D.Website: www.crypto.wustl.edu |
 |
Mark Johnston, Ph.D.Website: http://www.ucdenver.edu/...faculty/JohnstonM/Pages/JohnstonM.aspx |
Relevant Publications:
Kuttykrishnan, S., Sabina, J., Langton, L. L., Johnston, M., & Brent, M. R. (2010). A quantitative model of glucose signaling in yeast reveals an incoherent feed forward loop leading to a specific, transient pulse of transcription. Proc. Natl. Acad. Sci., 107(38), 16743-16748. doi:10.1073/pnas.0912483107
Funding:
 |
National Institute of General Medical ScienceWebsite: http://www.nigms.nih.gov/ |