


The Brent Lab is developing and applying computational methods for mapping gene regulation networks, modeling them quantitatively, and engineering new behaviors into them.
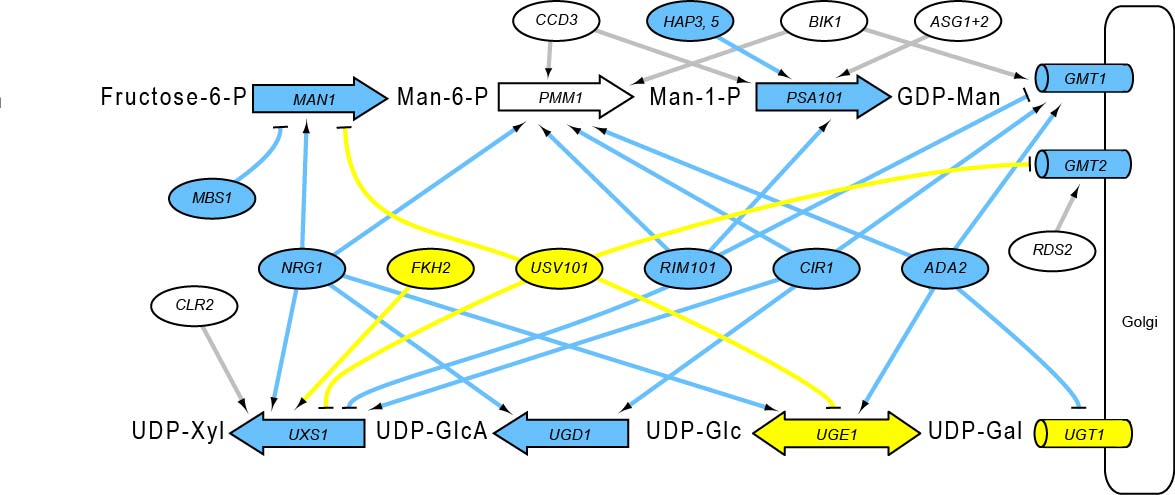
We develop efficient methods for determining which transcription factors can bind and regulate each gene in a genome.

We develop efficient algorithms for predictive modeling of transcriptional profiles as a function of the activity levels of transcription factors.
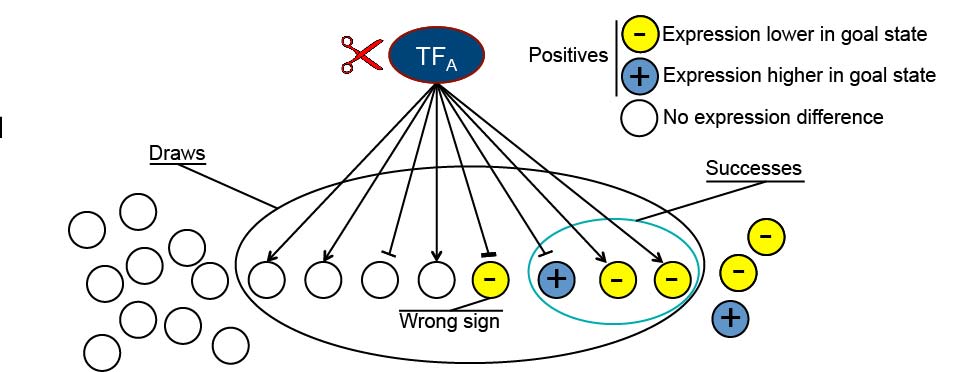
We develop algorithms for designing modifications to a transcription factor network that will drive the cell into a transcriptional state associated with a desired behavior.
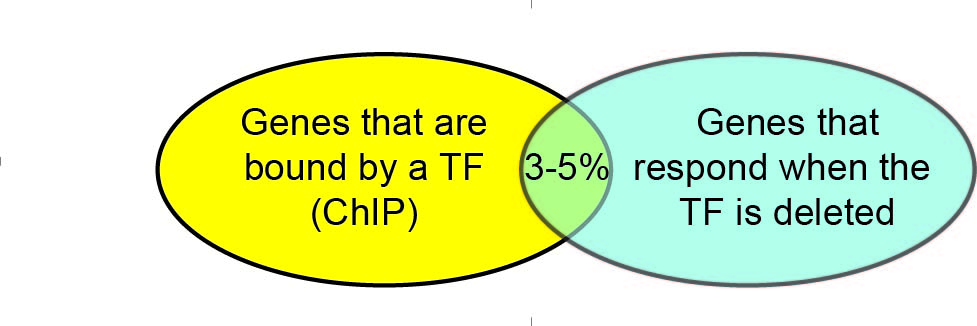
We are working to understand why some genes that are bound by a transcription factor are functionally regulated by that factor while others are not.
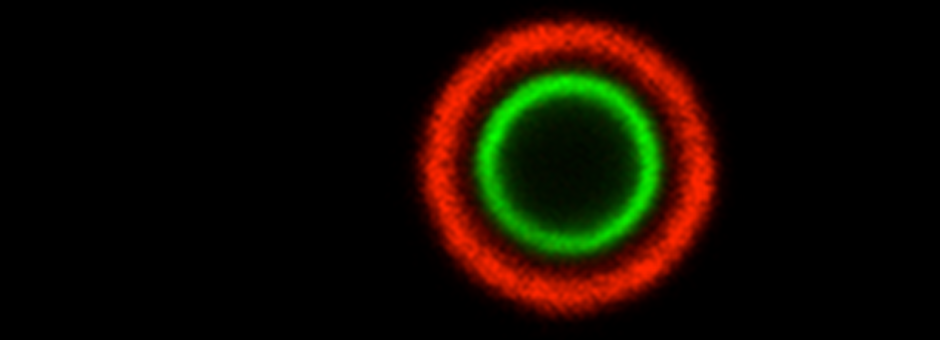
We use a powerful combination of molecular and computational techniques to reconstruct the regulatory network that controls C. neoformans capsule synthesis - in order to target this process in antifungal therapy.