Research
Overview
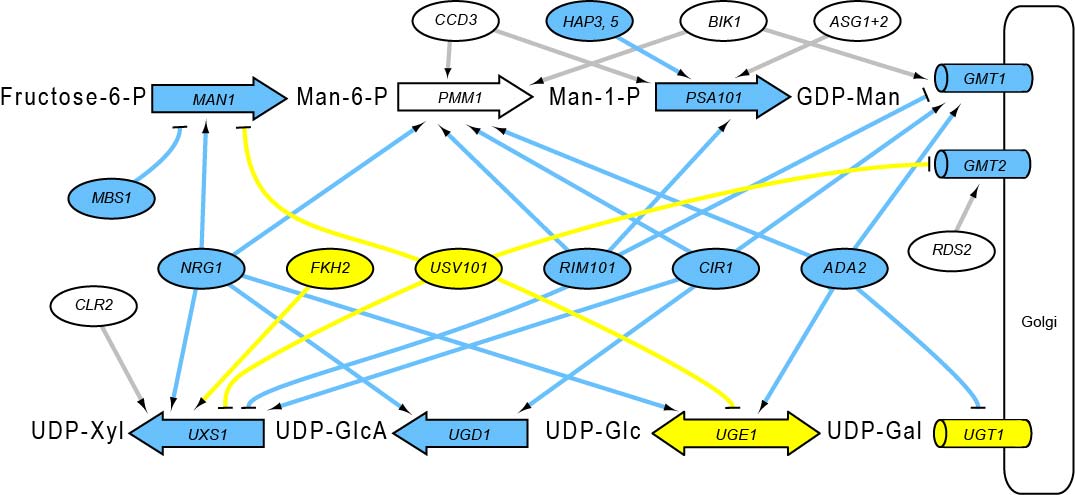
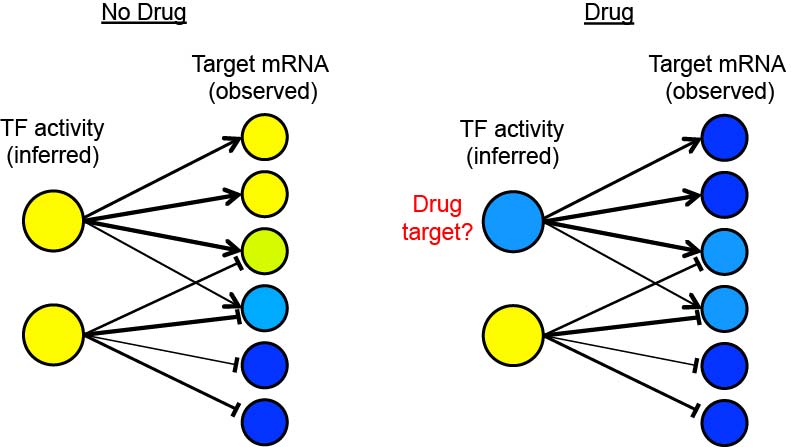
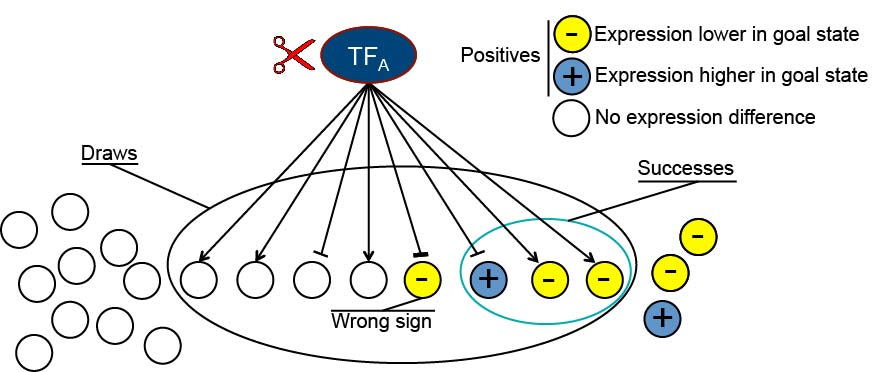
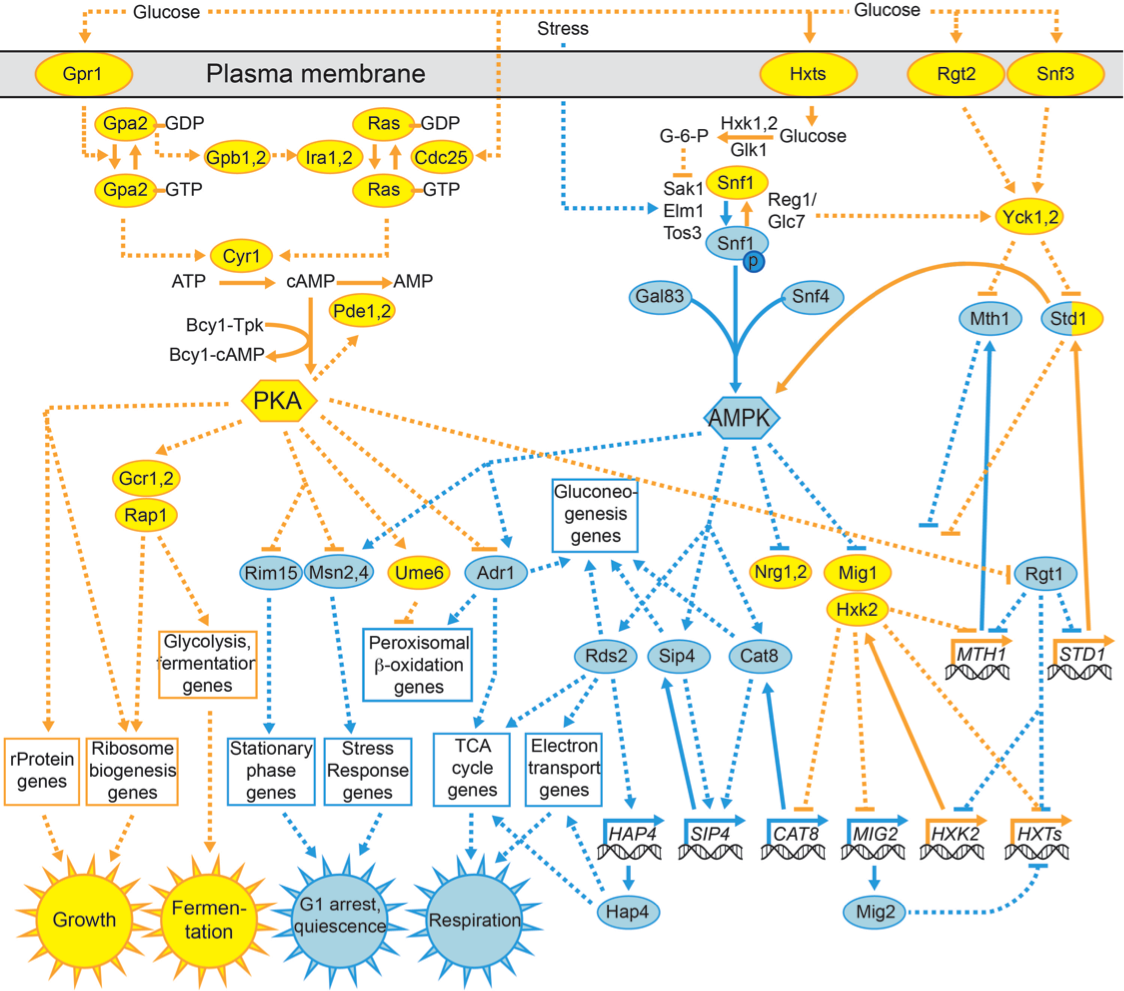
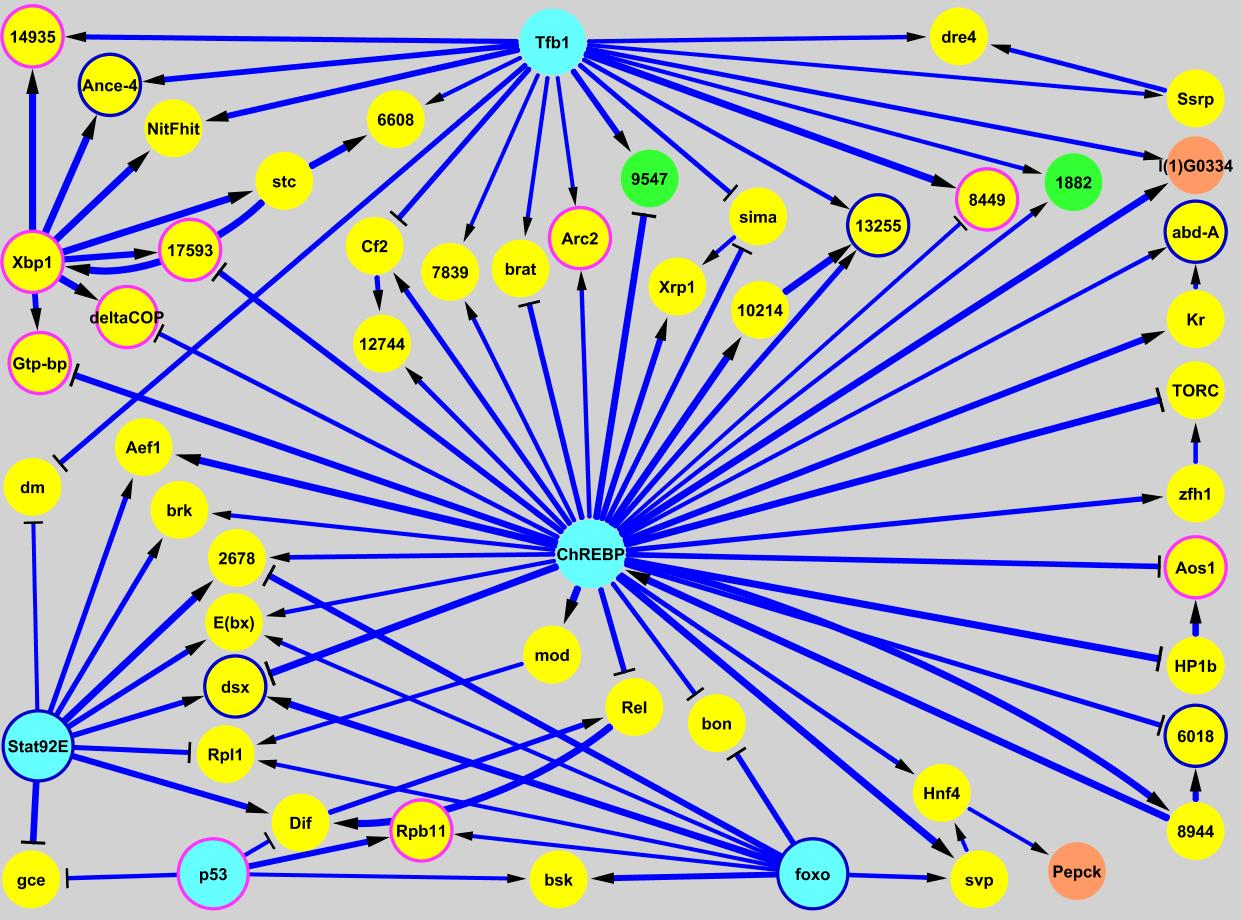
The Brent Lab is developing and applying mathematical/computational methods for mapping gene regulation networks, modeling them quantitatively, and synthesizing new network designs in living cells. We focus on scalable, "data light" methods that can be applied to a new organism from scratch within the scope of a single research lab, including data generation and analysis. The requirement to use only data generation that can be generated by efficient, predictable experiments is a significant constraint on the algorithms we develop. Our major projects are described below.