Research
Mapping transcription factor networks
Mapping a transcription factor (TF) network means determining which TFs have the potential to regulate each gene, given conditions that activate the TF. In 2013, we published the NetProphet algorithm for mapping TF networks by using gene expression data from cells in which a single TF has been genetically deleted. That paper demonstrated that NetProphet was the best available algorithm for mapping TF networks from gene expression data. We have now developed a second generation algorithm that significantly improves accuracy by using the amino acid sequences of the TFs and the DNA sequence of the gene's promoters. These algorithms have been shown to work well on yeast and fly TF networks; the next step is to optimize them for mammalian networks, which are much more challenging.
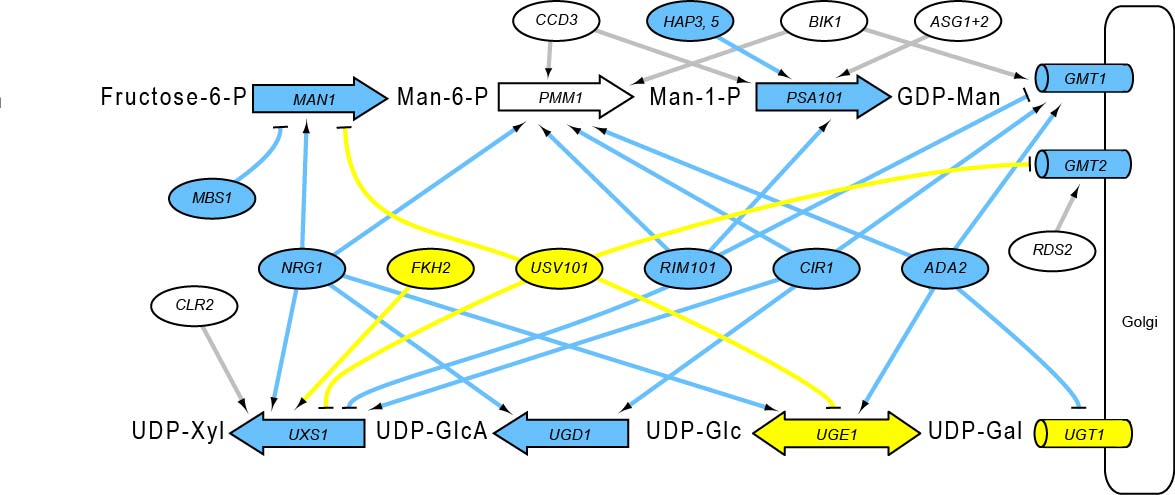
Regulatory relationships between TFs and the pathways that make and localize sugar donors for capsule synthesis in Cryptococcus neoformancs, considering the top 10,000 NetProphet edges: (large labeled arrows) synthetic steps; (ovals) TFs; (cylinders) nucleotide sugar transporters in the Golgi membrane. Shapes are labeled with the corresponding gene name and filled blue if the mutants are hypocapsular, yellow if the mutants are hypercapsular, and white if the gene has not been deleted (PMM1) or the mutants have normal capsule thickness (all others). Arrowheads indicate activation and T-heads repression; edge colors reflect the phenotype of the regulator. (Man) mannose; (Xyl) xylose; (GlcA) glucuronic acid; (Glc) glucose; (Man1) phosphomannose isomerase; (Pmm1) phosphomannomutase; (Psa1) GDP-mannose pyrophosphorylase; (Uxs1) UDP-Xyl synthase; (Ugd1) UDP-Glc dehydrogenase; (Uge1) UDP-Glc epimerase.
Current Students:
 |
Yiming KangEmail: yiming.kang[at]go.wustl.edu |