Research
Modeling Dynamic Transciption Factor Networks
Cells regulate their genes by changing the activity levels of transcription factors (TFs) in response to external conditions or signals. TF activity levels are an essential component the state of the network in a given condition, but they cannot be measured directly. Instead, they must be inferred from the amount of RNA being made from each of their target genes. TF activity (TFA) inference uses a static network map (see TF Network Mapping), which represents the potential of each TF to regulate each gene, together with RNA measurements in a variety of conditions, to infer how much of that regulatory potential is operative in a given condition.
TF activity inference can be cast in the framework of matrix factorization, in which the matrix of observed expression levels of genes across a variety of conditions is factored into a product of two matrices that need to be inferred. The first, called the control strength matrix, specifies how strongly each TF affects each of its target genes when it is active. The second, called the TF activity matrix, specifies the relative activity level of each TF in each condition. So far, this has not been applicable to genome-wide networks because the computation time required to carry out the inference/optimization procedures was too slow. We are developing faster algorithms and applying them to both the regulation of metabolism in yeast and the regulation of capsule growth in Cryptococcus. We aim to demonstrate the usefulness of TF activity inference for understanding biology and to make it a standard, widely used tool for analyzing cellular state.
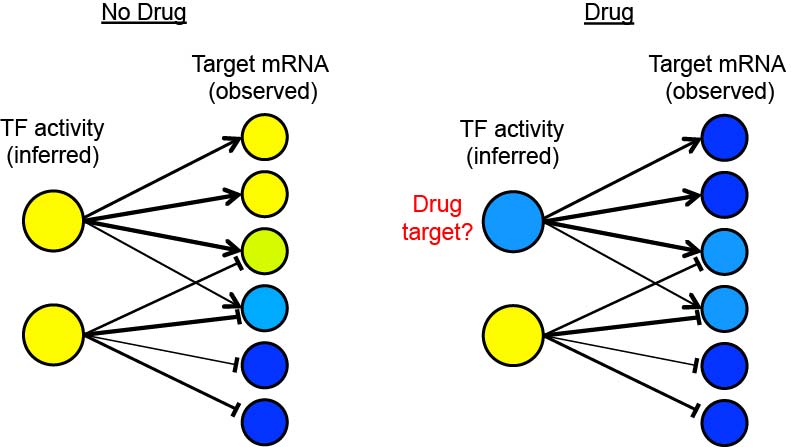
Transcription factor (TF) activity inference from qualitative network maps and condition-specific expression profiles. Large circles: TFs. Small circles: target genes. Line thickness: strength of regulatory influence. Arrow head: activation judi togel. T head: repression. Blue: low observed expression level (targets) or inferred activity level (TFs). Yellow: high observed expression level (targets) or inferred activity level (TFs).
Current Students:
 |
Cynthia MaEmail: czma[at]wustl.edu |
Relevant Publications:
Our lab has not yet published on this topic, but the basic idea was first laid out in:
Liao, JC, Boscolo, R, Yang, YL, Tran, LM, Sabatti, C, & Roychowdhury, VP. (2003). Network component analysis: reconstruction of regulatory signals in biological systems. Proc Natl Acad Sci U S A, 100(26), 15522-15527.