Research
Transcriptome Engineering
Transcriptome engineering means genetically modifying a cell's transcription factor network in order to change its transcriptional state (the amount of RNA present from each gene) to some desired objective. Transcriptome engineering is most frequently used to convert cells of one type (e.g. skin cells) in to another type (e.g. liver cells).
In 2016, we published
a paper on our transcriptome engineering algorithm, NetSurgeon,
in
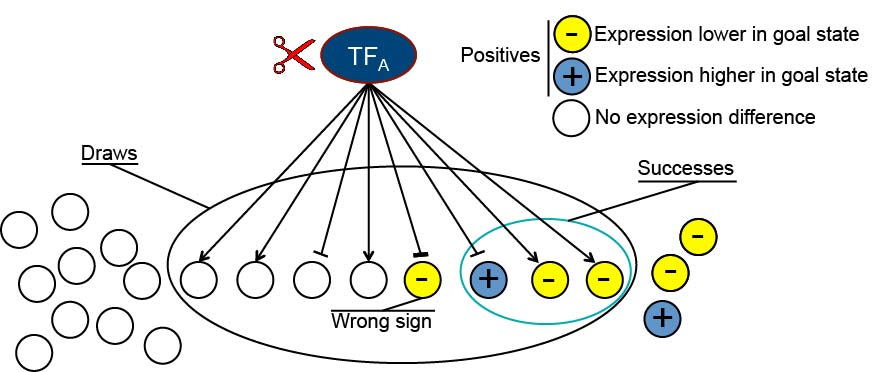
NetSurgeon scores are calculated using the hypergeometric probability function with draws, positives, and successes as indicated. Arrowheads indicate activation, and T heads indicate repression.
Current Students:
 |
Yiming KangEmail: yiming.kang[at]go.wustl.edu |